Scientific Computing
The Scientific Computing team supports researchers by performing numerical simulations that complement experimental research. In particular, we use state-of-the-art software to perform computational quantum mechanical modelling, molecular dynamics simulations, lattice dynamics calculation, data analysis and visualisations.
Software
Molecular Dynamics Simulations.
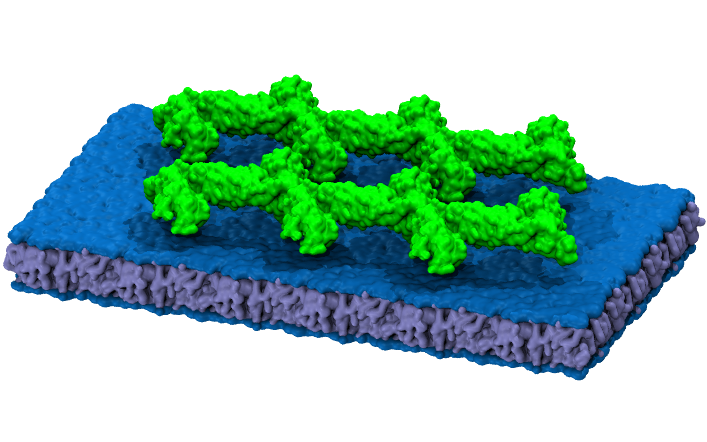
Applications of molecular dynamics simulations include the quantification of physical properties at the molecular level. In particular, MD simulations help characterise materials and study proteins and other complex molecules.
Molecular mechanics simulations use classical mechanics approximation in which the system's potential energy is calculated as a function of the nuclear coordinates using force fields.
With this kind of simulation, we can evolve systems of up to millions of atoms. However, this approximation does not fully describe the quantum state and interactions of the atoms. On the other hand, an optimised simulation of 100,000 particles can solve the system at a rate of 200 ns/day. Therefore, these simulations are suitable for the coarse description of relatively large systems and long simulations.

Image from: skblnw.github.io/mkvmd_render
Learn more aboutMolecular Dynamics Simulation Software
Quantum Dynamics Simulations.
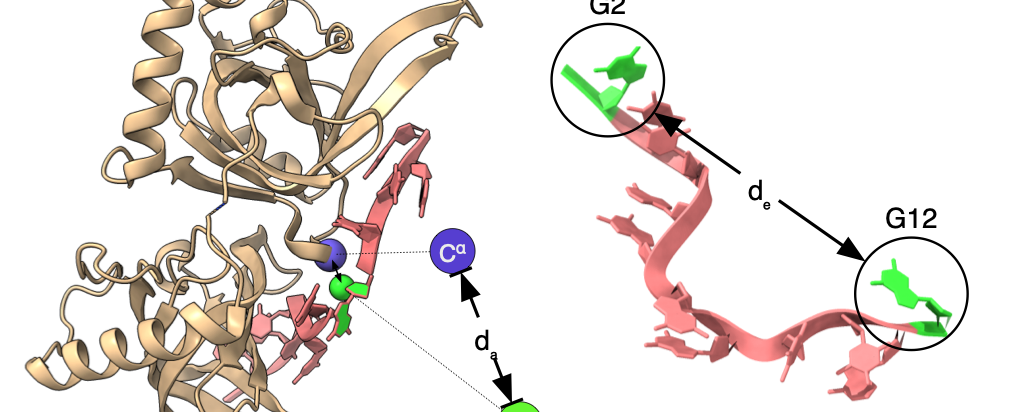
We can employ ab initio (or "from first principles") modelling software for experiments requiring detailed atomic-scale materials modelling. This type of software computes an approximate solution to the Schrödinger equation.
Ab initio calculations complement the experimental data with detailed information about the material at the molecular level.
Some of the calculations performed with this approach include ground-state calculations, structural optimisation, transition states, minimum energy paths, vibrational motions, magnetic spin-exchange parameters and magnetic moments, among many other quantities.
With this approach, it is possible to solve systems of up to a few thousand atoms for up to 10 ps. In addition, typical calculations of this kind provide a more accurate quantification of the system's state. For instance, the calculations performed using classical Molecular Dynamics employ a single-precision floating point. In contrast, Quantum Dynamics simulations are routinely run using double-precision floating-point.

Image from: APS / Alan Stonebraker
Learn more aboutQuantum Dynamics Simulation Software
Quantum Mechanics/Molecular Mechanics approach
Unfortunately, the Quantum Mechanics approach does not scale up well. In particular, the CPU time increases typically as the 3rd power with the number of bases set functions in DFT-based methods. Fortunately, combining the Quantum Mechanics and Molecular Mechanics approaches can produce more accurate and efficient simulations.
In QM/MM simulations, a small part of the system is treated quantum-mechanically. The remaining system is treated classically, thus allowing for studying chemical processes in solution and proteins or other complex interactions.
Applications of QM/MM include hybrid simulations of systems in systems where chemical reactions occur.
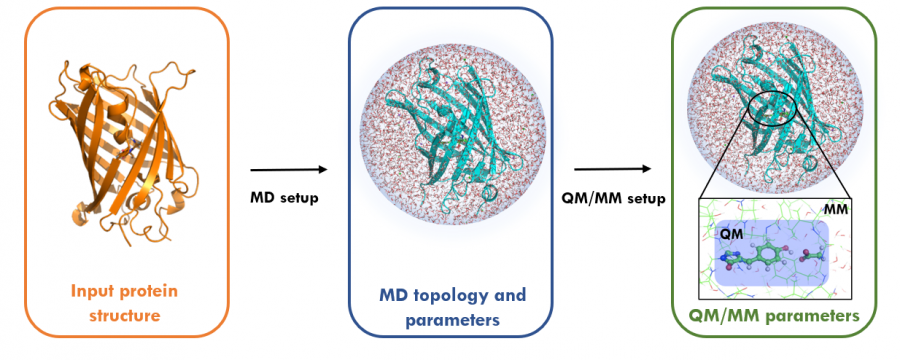
Image from: gromacs.org
Visualisation and data analysis.
Data analysis for simulations can be carried out with a variety of software packages, for example:
Visual Molecular Dynamics (VMD) is a molecular modelling and visualisation computer program.
UCSF Chimera is an extensible program for interactive visualisation and analysis of molecular structures, including density maps, supramolecular assemblies, sequence alignments, docking results, trajectories, and conformational ensembles.
MDANSE (Molecular Dynamics Analysis for Neutron Scattering Experiments) is a python application designed for computing properties that can be directly compared with neutron scattering experiments such as the coherent and incoherent intermediate scattering functions and their Fourier transforms.
Hardware
High-Performance Computing Cluster.
Atomistic simulations are computationally intensive. We need to solve complex equations describing the interactions between different components of molecules. Current computational technology allows us to solve relatively large systems efficiently.
We have an HPC cluster with 57 computing nodes (3458 cores) with a total memory RAM of 13954Gb and 3.1Pb of storage. To complement experimental results using atomistic simulations, we have six dedicated nodes. In addition, we have a node with 64 cores and 235Gb of RAM for visualisation, data analysis and interactive processing.
Contact us
Ask us a question.
